NetAD
Network modeling of bulk and single-cell multi-omics data to identify drivers for Alzheimer's Disease
Specific Aim of the Study
 The long-term goal of this project is to increase our understanding of Alzheimer's disease (AD) by harmonizing and modeling cross-study multi-omics data at the bulk and single-cell levels using network-based systems biology tools, thereby providing new therapeutic targets towards AD prevention and treatment. AD is the leading cause of dementia, affecting more than 5 million people in the US. Despite extensive investigation of AD pathogenesis and significant investment in developing therapeutics for AD, no cure is currently available. Characterization of AD brains by proteomics, transcriptomics, and genomics technologies has played a vital role and laid the foundation in dissecting the molecular bases of AD. However, few AD driver genes or proteins have been reliably identified. Therefore, sophisticated computational modeling tools and harmonized data visualization portals remain urgently needed to integrate bulk and single-cell multi-omics data across studies and platforms for the identification of "hidden" drivers for AD.
The long-term goal of this project is to increase our understanding of Alzheimer's disease (AD) by harmonizing and modeling cross-study multi-omics data at the bulk and single-cell levels using network-based systems biology tools, thereby providing new therapeutic targets towards AD prevention and treatment. AD is the leading cause of dementia, affecting more than 5 million people in the US. Despite extensive investigation of AD pathogenesis and significant investment in developing therapeutics for AD, no cure is currently available. Characterization of AD brains by proteomics, transcriptomics, and genomics technologies has played a vital role and laid the foundation in dissecting the molecular bases of AD. However, few AD driver genes or proteins have been reliably identified. Therefore, sophisticated computational modeling tools and harmonized data visualization portals remain urgently needed to integrate bulk and single-cell multi-omics data across studies and platforms for the identification of "hidden" drivers for AD.
To tackle this challenge, we propose a solution of network modeling of AD (NetAD). We have assembled a strong team with diverse expertise in systems biology (Yu), AD biology and proteomics (Peng, Wang, Li), statistical genomics (Wang), visualization (Zhou), and cloud engineering (McLeod), in collaboration with Drs. Haroutunian and Bu (AD pathology & pathogenesis). We have pioneered the development of network modeling algorithms (NetBID, scMINER) to integrate bulk and single-cell omics data and expose "hidden" drivers that may not be genetically altered or differentially expressed but drive disease phenotypes via post-translational modifications (PTMs) or others. We led AD proteomics studies through the development of our versatile JUMP software suite. We built the St. Jude Cloud platform for omics data sharing, analysis, and visualization. Capitalizing on our unique expertise and resources, we propose to implement NetAD with the following Specific Aims (Fig. 1).
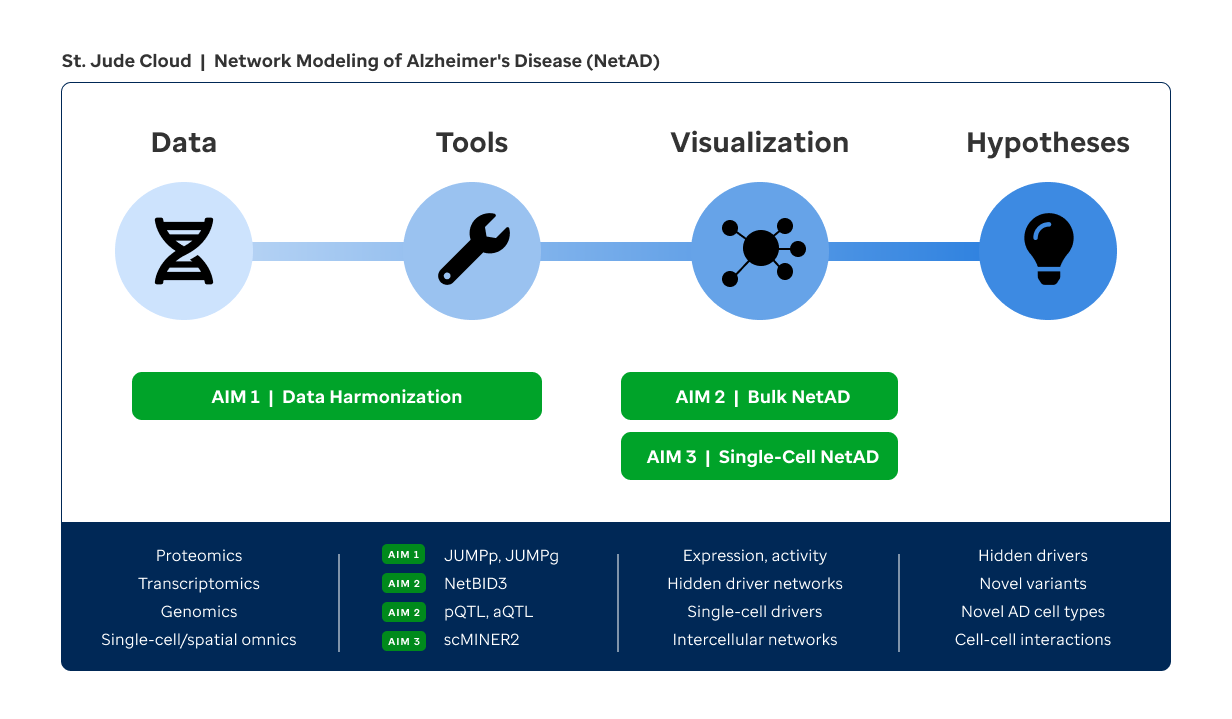

AIM 1
Develop sensitive and robust preprocessing, batch correction, and cloud-based visualization tools to harmonize cross-study transcriptomics, proteomics, and PTM-proteomics data of AD.
To address the challenge of harmonizing proteomics data, we will expand on our JUMP software to develop the JUMPp-lib algorithm for ultra-fast, sensitive identification of peptides using spectral library search optimized for isobaric labeling proteomics and the JUMPp-batch algorithm for imputing missing values and normalizing batch effects. We will then use the JUMP suite to quantify proteomes/PTM-proteomes (JUMPp) and genotypes (JUMPg) from 1,000+ TMT-based proteomics profiles. We will use our well-established in-house pipeline to reprocess 1,000+ RNA-seq profiles from multiple AD studies. Using our St. Jude Cloud platform, we will build a NetAD portal to centralize and freely share the harmonized data, including the gene and isoform expression of both RNA and protein levels, and PTM sites, via St. Jude Cloud Community, AD Knowledge Portal, and others.

AIM 2
Develop a network-based framework to integrate bulk transcriptomics, proteomics, PTM-proteomics, and genomics data to identify AD drivers.
Capitalizing the success of our NetBID algorithm, we will develop a new version (NetBID3) to infer AD hidden drivers by reverse-engineering a reference AD interactome from samples with matched transcriptome-proteome profiles and then using this AD map to infer peptide/PTM-peptide activity across all 1,000+ brain cases, many of which had only RNA-seq or proteomics data. We will extend our expression quantitative trait loci (eQTL) analysis to proteome/PTM-proteome (pQTL) and NetBID3-inferred protein activity (aQTL) to identify the underlying AD genetic drivers. Finally, we will develop tools in our NetAD portal to interrogate and visualize the interactome, driver activity, and pQTL/aQTL results.

AIM 3
Develop a single-cell network modeling algorithm to integrate single-cell and spatial omics datasets in humans and mice to identify cell-type-specific AD drivers.
Given the continuously increasing single-cell profiles of AD, we will develop a scMINER2 algorithm to integrate scRNA-seq data from human and mouse AD brains and model cell-type-specific networks and hidden drivers of AD. We will use scMINER2 to reconstruct ligand-receptor-driven cell-cell communication maps from single-cell data. We will also integrate available spatial transcriptomics data of AD brains to identify spatial patterns of NetBID3/scMINER2-identified AD hidden drivers and mapped cell types. We will develop visualizations for single-cell data in the NetAD portal.
This NetAD project will provide an innovative network modeling solution with new tools and a cloud-based easy-to-access portal to explore and visualize harmonized multi-omics data, networks, and drivers of AD. It will enable the generation of new hypotheses for "hidden" drivers of AD that may lead to novel AD therapies.
PIs:
- Jiyang Yu, PhD, Associate Member, Department of Computational Biology
- Junmin Peng, PhD, Member, Departments of Structural Biology and Developmental Neurobiology, Director of the Center for Proteomics and Metabolomics
- Xusheng Wang, PhD, Assistant Member, Department of Biology, University of North Dakota
Co-Investigators/collaborators:
- Xin Zhou, PhD, Assistant Member, Director of Data Visualization, Department of Computational Biology
- Yuxin Li, PhD, Principal Bioinformatics Research Scientist, Center for Proteomics and Metabolomics
- Clay McLeod, Director of Product Development and Engineering, Department of Computational Biology
- Vahram Haroutunian, PhD, Professor, Department of Psychiatry and Neuroscience; Director, The Mount Sinai NIH Brain and Tissue Repository (NBTR), The Mount Sinai School of Medicine
- Guojun Bu, PhD, Mary Lowell Leary Professor of Medicine; Chair, Department of Neuroscience; Associate Director, Alzheimer"s Disease Research Center, Mayo Clinic Jacksonville
Apps
Data from the NetAD study will be included in the following St. Jude Cloud applications.
Sequencing Data and Analysis
Genomics Platform
Next-generation sequencing data from NetAD will be available to analyze in the cloud.
Cite the Program
Please visit our dedicated documentation page on how to cite St. Jude Cloud when publishing manuscripts utilizing data or information from this resource.
Questions or Feedback? Contact us at support@stjude.cloud.