Welcome to the New St. Jude Cloud
April 15, 2020
Overview
We are excited to announce the new structure and accompanying redesign of St. Jude Cloud. Since the project's launch two years ago, the vision for St. Jude Cloud has steadily expanded from sharing our pediatric genomics data globally to developing a vibrant ecosystem of sharing at St. Jude Children's Research Hospital. By creating a scalable framework for increasingly large and diverse data types, our restructure supports new features today and lays the foundation for additional sharing resources to come.
What Has Changed?
The major shift in organizational structure is the transition from "Data, Tools, and Visualizations" towards a set of independent applications called apps. Each app gets its own identity within the St. Jude Cloud ecosystem and is designed to address a particular use-case. Apps are developed by teams across St. Jude and offer integrations between each other to enable cross-functional workflows. You can read the sections below to learn about the apps we offer today or view the map provided in our documentation to learn which app old functionality moved into.
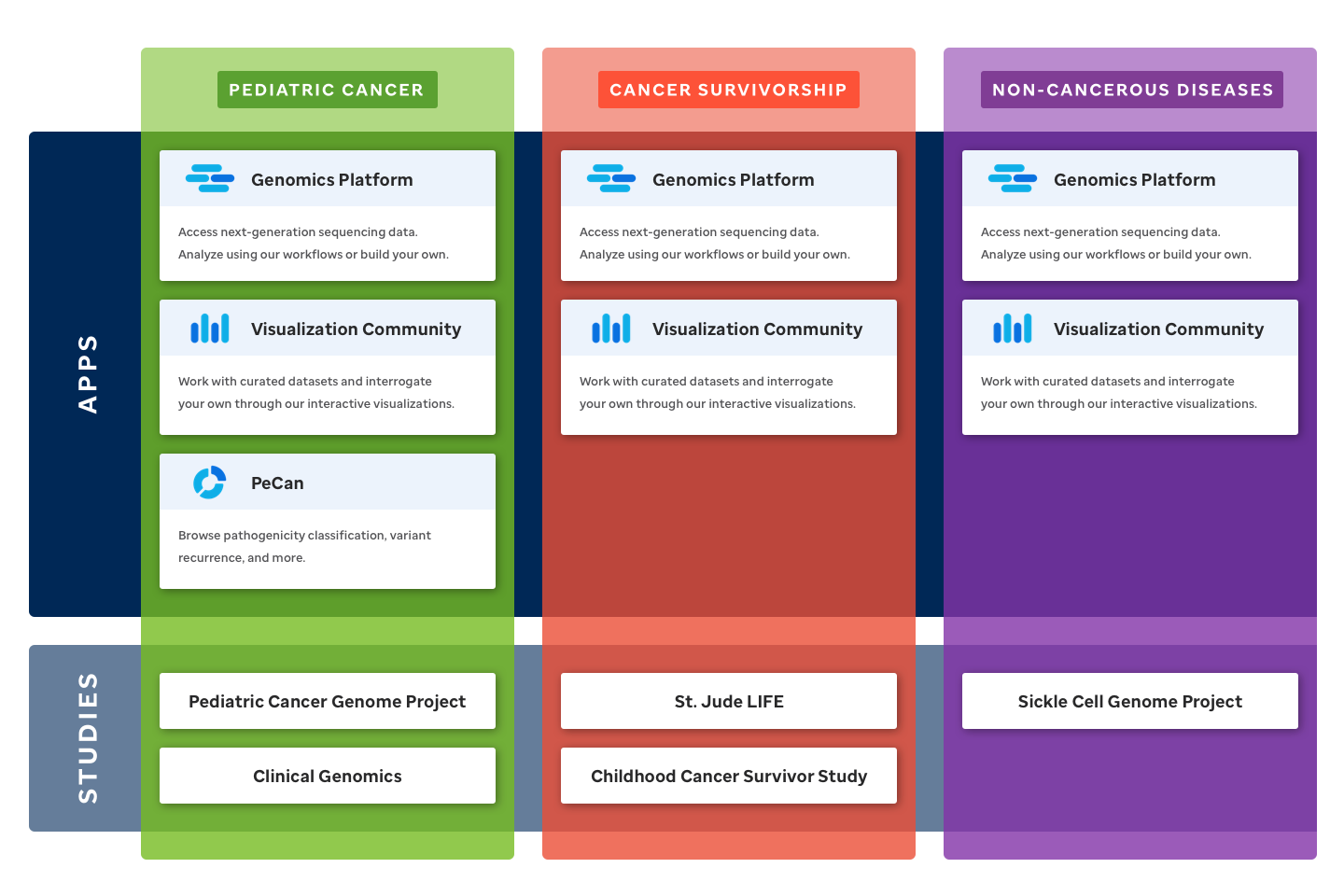
As the diversity of offerings in St. Jude Cloud grows, it is increasingly important for new users to be able to quickly understand which areas of St. Jude Cloud are relevant to their research. To address this, we’ve redesigned the home page around broad areas of study called research domains. These pages act as a signpost to areas of St. Jude Cloud that are relevant to researchers in these fields. All of this is accompanied by a refreshed look and feel that more accurately represents the project's unique mixture of technology and science. To learn more about how St. Jude Cloud can be leveraged in your research, view the domain pages for pediatric cancer, cancer survivorship, and non-cancerous diseases.
Genomics Platform

St. Jude Cloud Genomics Platform enables users to access and analyze large next-generation sequencing (NGS) datasets in the cloud. Built on top of the DNAnexus ecosystem and powered by Microsoft Azure, users can easily request access to the petabyte-scale NGS data in St. Jude Cloud alongside their own data to be processed by a wide variety of community-written analysis tools.
Features of the St. Jude Cloud Genomics Platform include
- Request access the world's largest repository of pediatric cancer related genomics data for analysis in the cloud or download.
- Upload your data alongside St. Jude's data to do cross-cohort analysis in the cloud.
- Leverage any of St. Jude Cloud's end-to-end computational workflows written by experts in the respective areas.
To get started, visit the Genomics Platform home page.
Pediatric Cancer Portal (PeCan)
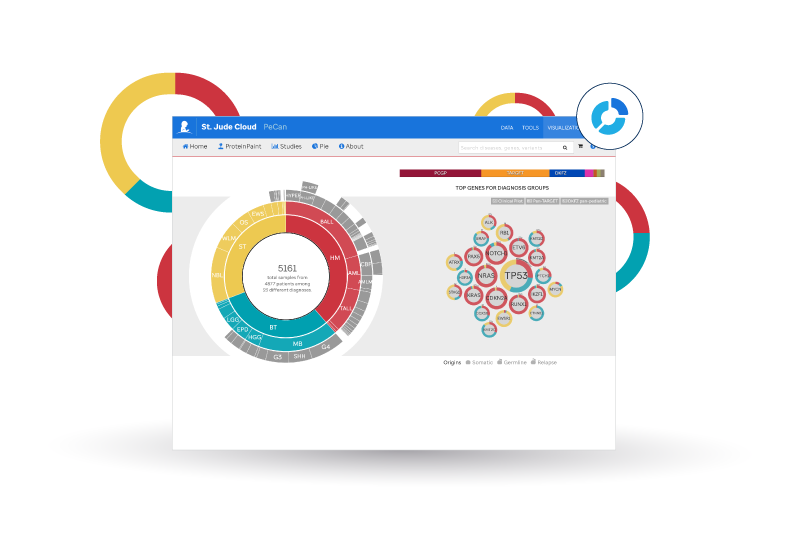
St. Jude Cloud PeCan features the pediatric cancer variation knowledge base curated by researchers at St. Jude Children's Research Hospital. This application presents a cohesive picture of the ever-growing knowledge we discover about pediatric cancer through analyzing omics data. You can browse information on pathogenicity classNameification, variant recurrence, and much more from samples contributed by St. Jude Children's Research Hospital, NCI TARGET, German Cancer Research Center, Shanghai Children's Medical Center, UT Southwestern, the Broad Institute, and the Medulloblastoma Advanced Genomics International Consortium (MAGIC).
Features of the St. Jude Cloud PeCan include
- Explore recurrently hit genes for subtypes of pediatric cancer using the donut chart on the home page.
- View mutation hotspots and gene expression data in pediatric and adult cancer. Click here for an example of viewing NRAS.
- Dedicated Variant Pages which describe in-silico triaging, population and variant frequencies, pathogenicity assessment by the St. Jude clinical genomics tumor board, relevant publications, and more. Click here to see an example for NRAS G12D.
- Run St. Jude's in-silico variant triaging algorithm against your variants using the PeCan Pathogenicity Information Exchange (PeCan PIE).
To get started, visit the PeCan home page.
Visualization Community

St. Jude Cloud Visualization Community, the newest addition to the St. Jude Cloud ecosystem, is home to St. Jude’s unique visualization tools such as ProteinPaint, GenomePaint, and SJCharts. Here, you can view community created visualizations using St. Jude tools, create visualizations with your own data which can be shared with collaborators and included in papers, and learn how to embed your visualizations in your website.
Features, both existing and those in development, include
- View community-created visualizations that use ProteinPaint, GenomePaint, and SJCharts.
- Create your own ProteinPaint, GenomePaint, and SJCharts visualizations (coming soon!).
- Host the data driving your visualizations securely in the St. Jude Cloud Genomics Platform (coming soon!).
To get started, visit the Visualization Community home page.
Conclusion
We feel confident that this new structure is a more scalable foundation on which we can expand the types of data shared on St. Jude Cloud. In fact, we're already planning the next set of features, including applications to explore rich, longitudinal data from our Childhood Solid Tumor Network and Neurobiology and Brain Tumor programs and a site-wide account that can be used to access all of the applications instead of an account per application. If you'd like to keep in the loop on these developments, we encourage you to subscribe to our email list using the link below the article.
For more detailed information about the restructure or how to use these apps, you can visit our documentation. With any feedback or questions not addressed by the documentation, you can contact our support team. For media inquiries, please contact our media relations team.